In the last century we have gleaned so much knowledge about ourselves physiologically. We have completed a tremendous project in 2003 called the Human Genome Project (HGP) in a quest to discover how our genes express disease. Francis Collins the director of the National Human Genome Research Institute stated in June 26 2000 :
“I would be willing to make a prediction that within 10 years we will have the potential of offering any of you the opportunity to find out what particular conditions you maybe at increased risk for, based upon the discovery of genes involved in common illnesses like diabetes,hypertension,heart disease and so on.”
A foolish statement since common illnesses such as diabetes 2, Hypertension and most heart disorders are lifestyle and dietary driven and certainly not a result of genetic anomalies.
Sixteen years on, new projects have taken root such as the Epigenome, and the Exposome to further our knowledge of gene expression and the microbiome or gut flora that resides in us all.
Scientists are hopeful that the completion of these new ventures coupled with the knowledge gathered from the Human genome project we will possess the total picture of how the human organism functions and manufacture more drugs and modalities with a greater success rate than what exists today.
I suppose some scientists believe that eventually we will know what God knows or has designed.
The Purpose of the Human Genome Project
The main purpose of the human genome project was to sequence or map the 3 billion DNA base pairs to identify, what we now know, the 20-25,000 genes that populate our 46 chromosomes. In Feb 2001 when the majority of the genome was published Francis Collins stated that:
“It’s a history book – a narrative of the journey of our species through time. It’s a shop manual, with an incredibly detailed blueprint for building every human cell. And it’s a transformative textbook of medicine, with insights that will give health care providers immense new powers to treat, prevent and cure disease.”
It was evident in the minds of most scientists that were involved in this monumental task, that, once complete it would turn the tide of disease prevention and treatment, and advance our knowledge of medicine to a level not witnessed within the last 100 years.
Furthermore, as stated by Francis Collins one of goals of the human genome project was to identify genes that specifically ignited human disease so as pharmaceutical research could manufacture chemical substances to cure disease by altering gene expression.
Another foolish statement proposing that synthetic chemical substances have the power to cure disease; they never have and they never will.
The Discovery of Epigenetics
Genome scientists for many years conducting DNA research noticed that the gene clusters located within the 46 chromosomes were separated by stretches of DNA that were not used for coding for cell reproduction and were thought to be superfluous so it was thrown away.
Unbeknown to them the so called ‘junk‘ was in fact huge amounts of regulatory proteins used in epigenetic gene control (biologists label the coding part of DNA Exons and the epigenetic switching part Introns).
Epigenetic gene expression is far more complicated than just simply reading a fixed blueprint ( DNA) for cell reproduction.
For 50 years scientists believed (and is still in some biology textbooks) that the genes we inherited were fixed in stone and that we all were a ‘victim of our genes’ that we had no control over.
If dad had prostate cancer then there is a good chance that his son would inherit the gene that expresses this condition which is why human ‘disease’ was thought to be by and large genetic.
Even now most doctors still believe this to be true. It followed that the majority of earth’s population were led down the road of genetics being the root of most bodily ailments.
We now know that our genome has 3.2 billion bases (Our genes are DNA made from chemical substances Adenine, Cytosine, Guanine and Thymine A,C,G,T, since DNA is a double helix, A can only pair with T, and C with G which are base pair nucleotides), where the average size gene is 3000 bases and the largest gene measured to be 2.4 million bases is Dystrophin (a gene that makes a protein for muscle integrity found in skeletal muscle and the heart ) and Chromosome 1 houses the most genes = 4000+.
As you can see, this gets quite complicated, but the point is:
Only 3% of DNA is dedicated to gene division, the remaining 97% is gene regulation ( from the so-called ‘Junk DNA).
Have they discovered what gene expresses our common diseases like diabetes, hypertension, high cholesterol? Of course not because these are all symptoms of other underlying problems.
Do certain genes change with these ailments? Yes, genes respond to exercising, ingesting food, taking prescription drugs, breathing or lying in the sun ( e.g: the absorption of Vitamin D3 affects over 2000 genes ).
The Birth of Pharmacogenomics
A new ‘religion’ was born from the Human Genome Project called Pharmacogenomics. This involves more personalised medicine.
Once prescription drugs are metabolised by the liver, the recipient’s response is determined partially by their specific gene states ( expressed (ON ) or silenced (OFF)), so with this understanding, physicians can determine specific drugs and dosages on an individual level.
The ultimate goal is to develop genetic tests to predict how patients are going to respond. Coupled with this testing tool it is envisaged that side effects can be minimised.
The HGP identified changes in the nucleotide bases ( A,C,G,T) referred to as SNP (Single Nucleotide Polymorphisms) between individuals, and structural changes in various DNA sections, altering chromosomes (affecting some 12% of the genome), including translocation ( genetic material is exchanged between chromosomes).
Scientists realised that it’s difficult to ascertain which part of the SNP cause disease and which do not, except through statistical analyses between a number of individuals.
They’re looking not only for genes but gene variants that cause disease. This also can involve multiple genes and multiple variants; a bit like trying to hit a moving target.
Understanding Epigenetics
Several years after the completion of the Human genome project the Epigenome project was born.
Scientists realised that apart from identifying the human genes and their chromosomal location, disease expression was not simply caused by a hard wired DNA blueprint being read, thus creating a gene expression, but a more intelligent functioning was at play.
About the same time the Epigenome project was launched. Dr. Christopher Wild, a cancer epidemiologist, published a 2005 article entitled “Complementing the Genome with an “Exposome” which proposed that environmental factors had a role to play in human disease as if this was something new.
Hippocrates himself in 300BC examined the relationship between disease and environmental influence. Since scientists accepted that epigenetic gene switching was influenced by internal and external ( environmental) forces the Exposome sought to define these influences.
To understand Epigenetics, it is necessary to appreciate some basic biology.
Humans are made up of trillions of Eukaryotic cells, cells that contain a nucleus, where the DNA and chromosomes reside. Each cell contains approximately 2 metres of DNA and the typical human cell nucleus is 6 micrometers in diameter, so the DNA is supercoiled around scaffolding alkalised protein molecules called Histones (This is geometrically equivalent to packing 40 km of extremely fine thread into a tennis ball). This structure is known as a Nucleosome.
So a typical cell contains 6.4 billion base pairs which equates to 3.2 million nucleosomes.
The total collection of Nucleosomes make a Chromosome.
For example, Chromosome 1 is the largest chromosome in the Human, spanning 249 million nucleotide base pairs representing around 9% of the total DNA in the cell and is estimated to have 4300 genes such as Gene AHCTF1 which transcribes a protein called ELYS which is used to regulate the transport of molecules between the cell nucleus and the Cytoplasm ( a gel like substance that maintains cell integrity and is the home of enzymes that are responsible for cell waste management), and cell division.
According to scientists some genes on Chromosome 1 when expressed, are responsible for diseases such as Alzheimers and breast cancer.
The readers of this article should be aware that whatever environmental stimuli is present at any given time will influence the body’s response to that stimuli and epigenetic gene expression will occur which happens minute by minute, hour by hour, year by year throughout life, regulated by the body’s master adaptation process called ‘Methylation’ which is activated millions of times every second. To believe that genes are responsible for expressing disease displays a profound ignorance of how living organisms actually function.
The following diagram shows histone structure within DNA:
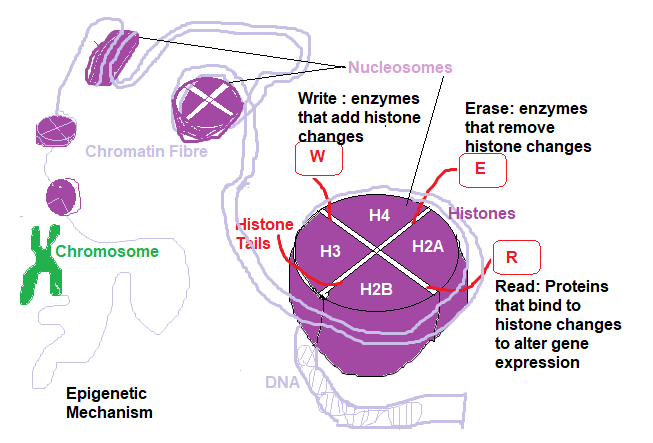
To regulate or control cell expression it is the Scaffolding Histone Proteins that are manipulated chemically (Chemical signalling). The histones are a cluster of base proteins referred to as H3,H4,H2A,H2B ( there are many more H3K4,H3K3,H3K9 etc dependent upon what Chemical signalling is used).
Each Histone has an N-Tail attached to it to which is activated by a chemical signal ( shown in the diagram providing Read, Write and Erase functions).
Histone function is complex, however in order for a section of DNA to be read it must be unwound from a particular histone cluster activated by a particular enzyme. This process involves adding a specific chemical group referred to as a Methylation,Acetylation, Phosphylation etc group.
These enzymes are Histone Methly Transferase ( HMT ) and Histone Acetyl Transferase (HAT) to unwrap the DNA section for Methylation and Acetylation respectively.
Enzymes to rewrap the DNA are Histone demethylase (HDMC) and Histone deacetylase (HDAC). Specifically, these enzymes convert the histones from unmethylated ( unacetylated, unphosphylated etc) to methylated histones.
Once the DNA section is uncovered and read , a copy is made called mRNA ( messenger RNA in this case). This action is referred to as Transcription. Once this is complete new or recycled histones are produced to rewind the DNA.
H4 is generally activated in this way using a acetyl group referred to as Acetylation. This Acetylation cycle, occurring within the cell nucleus, can go through multiple transcriptions, being modified several times before the mRNA produces the final protein sequence that is released into the cytoplasm from the cell nucleus. H3 is generally activated via an Methly group referred to as Methylation. However the type of methylation depends upon the amino acids ( in this case Lysine or arginine ) attached to its N-Tail. For example H3K3 ( H3+Lysine 3) expresses a gene as in the Methylation example but H3K9 leaves the gene unread ( thus silencing the gene ).
The Problems with Epigenetic Drugs
The Pharmaceutical industry is now having a field day developing epigenetic drugs. One of most researched diseases in relation to epigenetics is cancer. The targets include Inhibiting the enzyme (HDAC) that rewraps the DNA after reading leaving the DNA section lose and unwound which somehow triggers the transcription of a tumor suppression gene. Additionally DNA methylation inhibition is used which has the effect of silencing the cancer gene expression and somehow turns on tumor suppression.
One such drug that inhibits DNA methylation is Azacitidine which is used to treat myelodysplastic syndrome, a blood disorder that can lead to leukemia. However, like most drugs there are various possible side effects that may include:
- risk of infection due to low white blood cell count (immune system suppression)
- anaemia (possible need for blood transfusion)
- low platelet count (compromised blood clotting)
- constant fatigue
- diarrhea
- constipation
- loss of appetite
- chest infection
- dizziness
- loss of fertility
- sleep disorder
- Indigestion
- blood in the urine
- kidney malfunction
- liver dysfunction
- allergies
The human body thrives on equilibrium and once that balanced state is upset the body reacts and attempt to return to a balanced condition, however the ‘SNAFU’ effect that ensues from altering genes (short circuiting) abnormally, causes other associative processes to change triggering a flurry of ‘side effects’.
How Drugs affect our Microbiome
We mentioned Pharmacogenomics in an earlier paragraph that is designed to minimise side effects, but pharmaceutical companies need to get their drugs to market as quickly as possible, so they have neither the time nor the knowledge to minimise side effects.
Lupus a chronic autoimmune condition, where the immune system attacks host tissues, causing damage to joints, skin, kidneys, blood, the heart, and lungs, is, scientists claim, that they have uncovered aberrant histone modification associated with lupus like symptoms in mice. A drug called Trichostatin A is used to treat this condition which is also a HDAC inhibitor to correct acetylation at 2 histone sites. This drug is also used as an anti breast cancer drug.
Some of these drugs can also target Prokaryotic (Unicellular) cells that constitute life in bacterial organisms. The prokaryotic cells reproduce using the same mechanism of DNA to mRNA transcription, required for protein translation.
The difference between the Prokaryotic and the Eukaryotic cell is that the reproduction mechanism within the prokaryote occurs within the cytoplasm, since these cells have no nucleus, and multiple transcriptions and regulation steps do not occur and their non complexity ( in comparison to their Eukaryote counterparts) gives rise to fast rate proliferation.
Since the pharmaceutical industry can target both cells this begs the question,
When will our own microbiome be a target for these drugs?
Oh I forgot antibiotics destroys the microbiome. But the ‘Bugs to drugs’ initiative is near. Research has been in in progress for some time concerning Microbiome drug development.
“Could this be the final frontier?”
The Human Microbiome Project
In fact in 2008 the Human Microbiome project was launched more as a 5 year feasibility study. This study was under the leadership of Francis Collins.
In 2014, it was reported that our microbiome houses 10 times the amount of bacterial cells as there are human cells (100 trillion bacterial cells vs 10 trillion human cells), but we don’t really know, since other research proposes 37 Trillion cells constitute the human organism which would make it a 3:1 ratio as opposed to a 10:1 ratio).
The microbiome is made of mainly bacteria, single Prokaryotic celled microorganisms like yeast and parasitic worms (helminths) and viruses. The purpose of this project was to establish a documented reference of microbial genome sequences and explore the association of human disease and the gut flora.
Some 242 volunteers provided some 5,000 tissue samples for analysis and we now know that more than 10,000 microbial species occupy the human ecosystem and that more microbial genes contribute to the survival of a human than host genes.
However, not more than 150 microbial species occupy the average human at at any given time. I assume somewhere in the research an examination of the ‘cross talk’ between our gut flora and the human brain will occur. Anyway discussion on the microbiome and its true function in the human is reserved for future articles.
The common link between the human genome, epigenetics and the human microbiome initiatives is the focus of the exposome, namely environmental input.
This idea of mapping the human exposome appeared in 2005 and work has continued with analysing the 80,000 some industrial chemicals used in our everyday lives from breathing in a toxin to handling chemically made articles like credit cards and plastic bottles. Ingesting endocrine disruptor/antibiotic herbicides,pesticides, the food we consume, the prescription drugs that we take all have a profound effect on our lives.
The department of defence has has over 100 million blood samples from soldiers collected first, upon enlistment, and then after specific tours of duty. Various studies going back decades such as the child health study conducted by the University of California has collected millions of blood samples over a time span of 50 years looking at pesticide exposure from intensive farming.
Some Very Important Questions to be Answered
In summary, having completed the human genome project, and the on-going research into the human epigenome, the human microbiome and the human exposome, have we advanced toward reducing the number of health related deaths or decreased preventable adverse health conditions (Dis-eases) ??.
Has our health care programs improved as a result of this vast knowledge?
Have we eradicated some diseases that have plagued the human for the last 50 or 100 years specifically related to information collected from these ‘ome’ projects?
The simple answer is No, why ?….‘Stay Tuned’.

“It’s a dangerous business, Frodo, going out your door. You step onto the road, and if you don’t keep your feet, there’s no knowing where you might be swept off to.”
Lord of the Rings
Author: Eric Malouin
Reviewed by Dr. John Bergman D.C.